Let's be honest. Optimizing a purification protocol for a single peptide is not something that one wants to spend a lot of time thinking about, let alone actually pursuing. There are times though, that a little bit of effort in this arena can very impactful when considering the time required to fully synthesize, purify and deliver a peptide.
In today's post, I'll discuss an approach combining the use of a focused gradient with a step gradient that minimizes the steps necessary to identify a more optimal flash purification gradient for peptide purification.
Over the last few years, I have discussed several strategies that can help you transition from HPLC to High Performance Reversed-Phase Flash Chromatography (RP-HPFC) for peptide purification. Some of those strategies, including using a focused gradient or even a step gradient to improve the purity that you are able to achieve using RP-HPFC while still maintaining high recovery levels of the desired peptide. These two strategies do require some additional effort when compared to the "standard" gradient template to actually achieve higher purity though.
While a focused gradient can be identified usually with one or two injections on your flash system, a step gradient can often take numerous injections - one purification I worked on required 11 injections before settling on final optimal conditions - requiring a significant amount of time spread over multiple days.
Given the fact that the Biotage® Isolera™ Dalton 2000 takes information from a linear gradient, of any slope, to build the step gradient, I asked the question "will combining these two strategies help to minimize my method optimization time?" So I took a peptide sample I had laying around, confirmed the significant masses present in sample using the direct inject feature, and proceeded with a purification optimization.
Just a note here - the desired peptide mass was not present in this sample. However, the main product mass is suggestive of only a single amino acid missing from the desired peptide, which shouldn't be a significant change chromatographically. With this information in hand, I decided to proceed with the template-style linear gradient I would use for the intact peptide running from 20% to 70% MeCN over 10 column volumes (CVs), Figure 1.
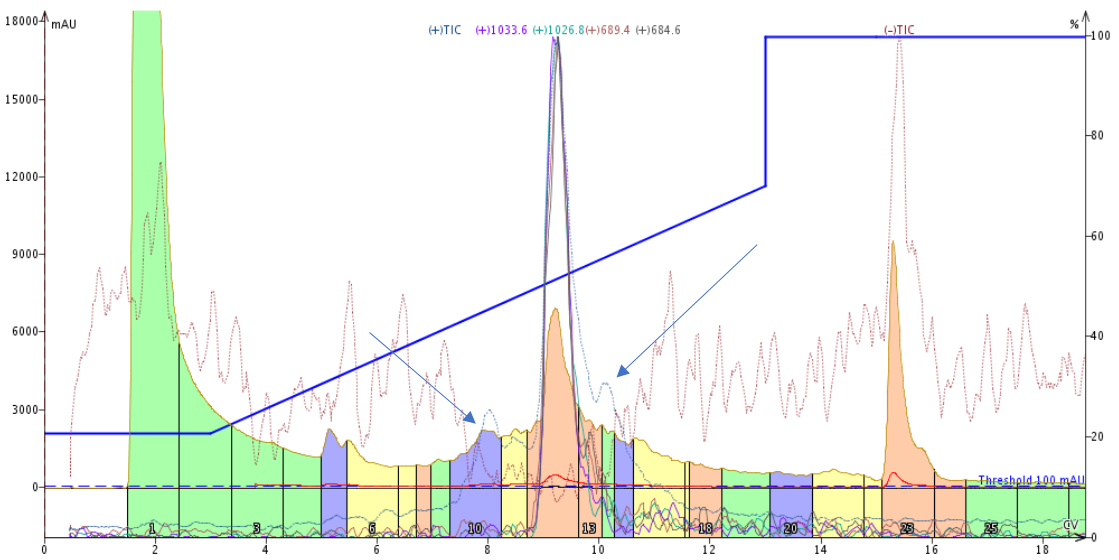
Figure 1. Peptide purification using a template-based linear gradient. While this separation appears pretty good, the positive TIC (light blue line) indicates the presence of two other significant masses (blue arrows) eluting near the desired product peak. The target product has a mass of 1033.6 Da as identified during the direction injection described above.
This purification performed about as expected. The major peak elutes midway through the gradient and a typical leading impurity is somewhat resolved. If we only look at the UV traces, it appears as though the product peak tails as it elutes from the column, but the Total Ion Chromatogram (TIC, blue arrows) indicates the presence of an additional compound in the product peak tail. Most importantly though, this templated gradient purification can certainly be improved with a focused gradient and likely further improve with a step gradient.
Let's first start with a focused gradient. According to the above purification, the major peak begins to elute at about 50% MeCN. We know that this percentage is a little bit high compared to reality inside the column, so I decided to start the focused region at 30% MeCN and increase MeCN concentration to 40% over 10 column volumes, Figure 2.
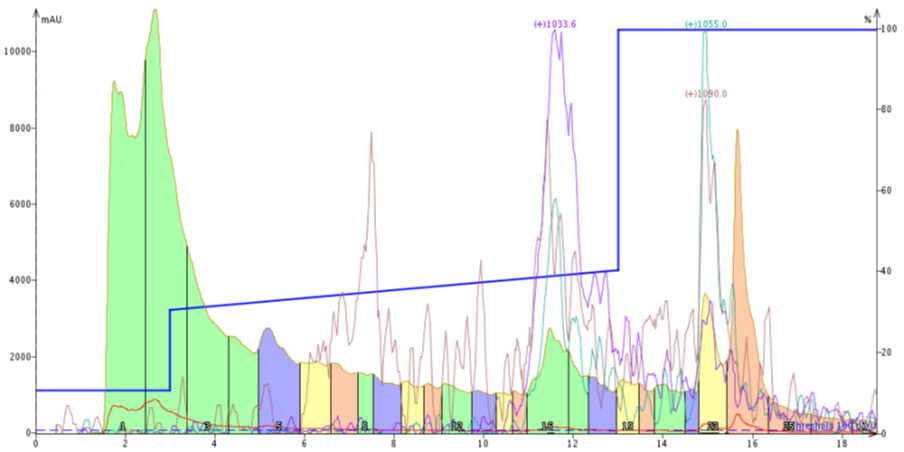
Figure 2. Peptide purification using a focused gradient. The gradient, 30% - 40% MeCN over 10 column volumes, was started slightly too low to ensure an improvement in purification.
Well.... based on this purification I think I need to try again. The desired compound elutes a little too late within the focused gradient for me to feel comfortable that a separation is actually occurring. There is also a distinct absence of the previously-observed leading impurity.
For the second focused gradient, I simply adjusted the starting and ending points slightly. In this case, I used a 35% - 45% over 10 CV, Figure 3.
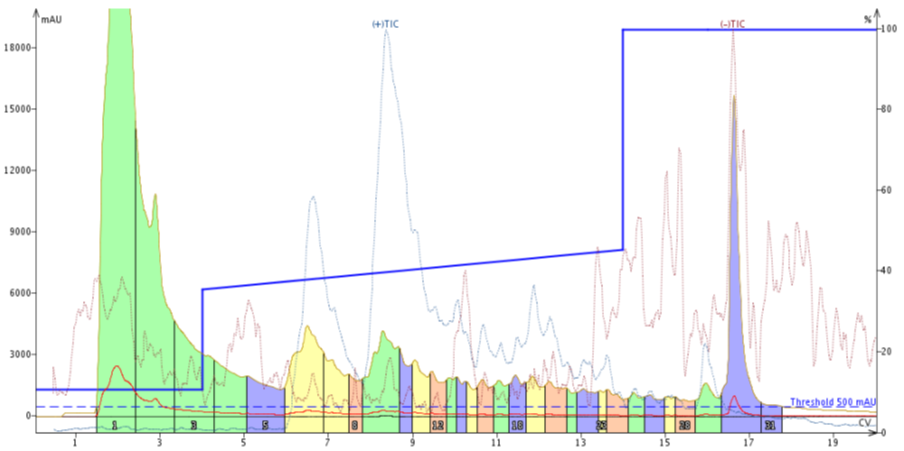
Figure 3. Peptide purification with an adjusted focused gradient. The gradient, 35% - 45% MeCN over 10 column volumes shows a dramatic improvement in separation compared to the previous attempt.
This is more like what I was expecting to see. The adjusted focused gradient enables improved resolution between the leading impurity and the target peptide. We also see improvement in resolution between the target peptide and the trailing impurity (more clearly identified by the TIC peak at about 12 CV). However, the TIC suggests yet again that there are additional compounds tailing out of the desired product peak.
With this improved linear gradient in hand I used the Optimize software feature on the Isolera™ Dalton 2000 to design a step gradient, Figure 4.
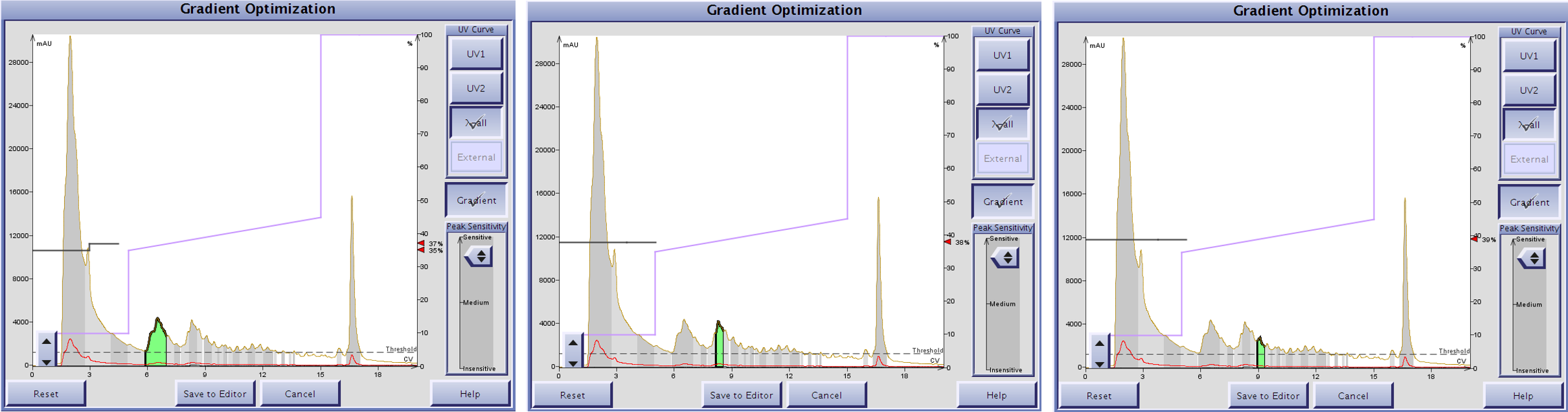
Figure 4. The Optimize software feature included with an Isolera Dalton 2000 predicts the MeCN concentration required to elute the selected peak (green) based on a linear gradient. Using the more optimal focused gradient as a reference, the software predicts MeCN concentrations of 37% (left), 38% (middle) and 39% (right).
One thing I have found when using the Optimize feature is that usually the final gradient has steps with slightly lower concentrations of MeCN than what is predicted. Using this knowledge, I built a step gradient starts at 10% MeCN to ensure compound retention by the stationary phase, then jumps up to 35% MeCN for 5 CVs to elute the leading impurity, then steps up to 37% MeCN for 5 CV to elute the compound of interest. From here, I stepped straight up to 100% MeCN to flush the column as the trailing impurity should still be retained, Figure 5.
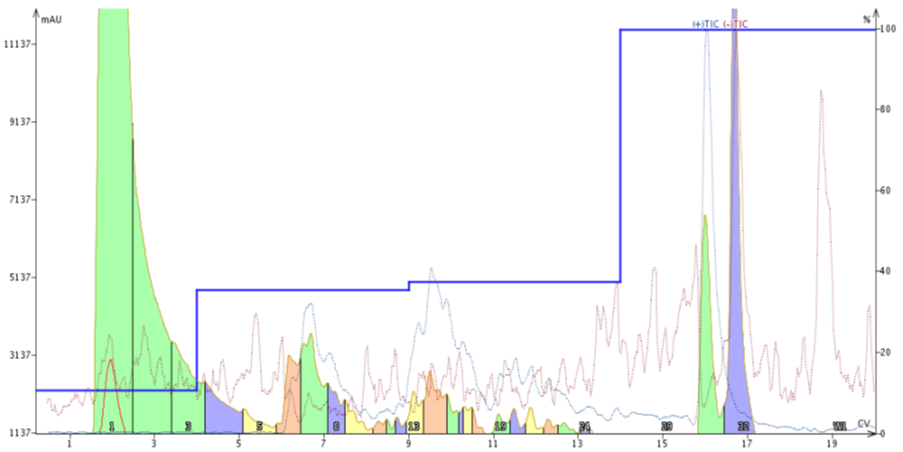
Figure 5. Peptide purification using a step gradient. The MeCN concentrations selected for the step gradient were guided by the Optimize software feature which used an improved focused gradient as reference.
This worked extremely well! The leading impurity is essentially baseline resolved (even with the TIC trace) and the previously coeluting impurity is also resolved. I haven't analyzed the main product fraction (middle, orange), but the mass analysis suggests that the desired fraction is highly pure.
What's most important to me in this process is just how quickly I was able to go from a mediocre, but templated purification to something that will most certainly yield a highly pure compound. The last time I used a step gradient for a difficult purification, I evaluated 10 different gradient conditions before I finally settled on something that fully resolves a 19 amino acid impurity from the 20 amino acid product. This process required only 4 injections to get to that same optimal point. In all reality, if I had thought a little harder a longer about my focused gradient, it could have reduced to three injections. Each of these injections requires only about 10 minutes to complete, and the additional MS information reduces the need for analytical HPLC between every injection.
If you would like to learn more about creating a focused gradient, click the link below to find a more detailed how-to guide.

