Synthesizing libraries of peptides - potentially hundreds in a single synthesizer setup - has seen a resurgence of late. These synthetic libraries, often prepared on very small synthesis scales, are required most recently for secondary screening preliminary structure-activity relationship determinations, or neoantigen vaccine preparations. However, optimizing the synthesis of hundreds of peptides to be made simultaneously is a very different beast when compared to optimizing the synthesis of a single peptide.
In today's post, I'll describe a strategy that can increase the overall crude purity of an entire peptide library using two different coupling chemistries.
Given the expansion of peptides in therapeutics, and specifically the need for synthesis of libraries of peptides, I decided it was time for me to start experimenting in this area. But peptide synthesis, when considering simultaneous synthesis of hundreds of compounds that may or may not be similar was pretty daunting the first time. After all, there is no longer one peptide to be concerned about (crude purity and yield predominantly), I now have to be worried about many many peptides. And ideally, the crude purity of the entire library would be sufficient that these could be take forward into validation experiments without further purification.
So I decided to start with something a little more "simple" - a scanning library centered around a peptide with which I already have some familiarity, Figure 1.
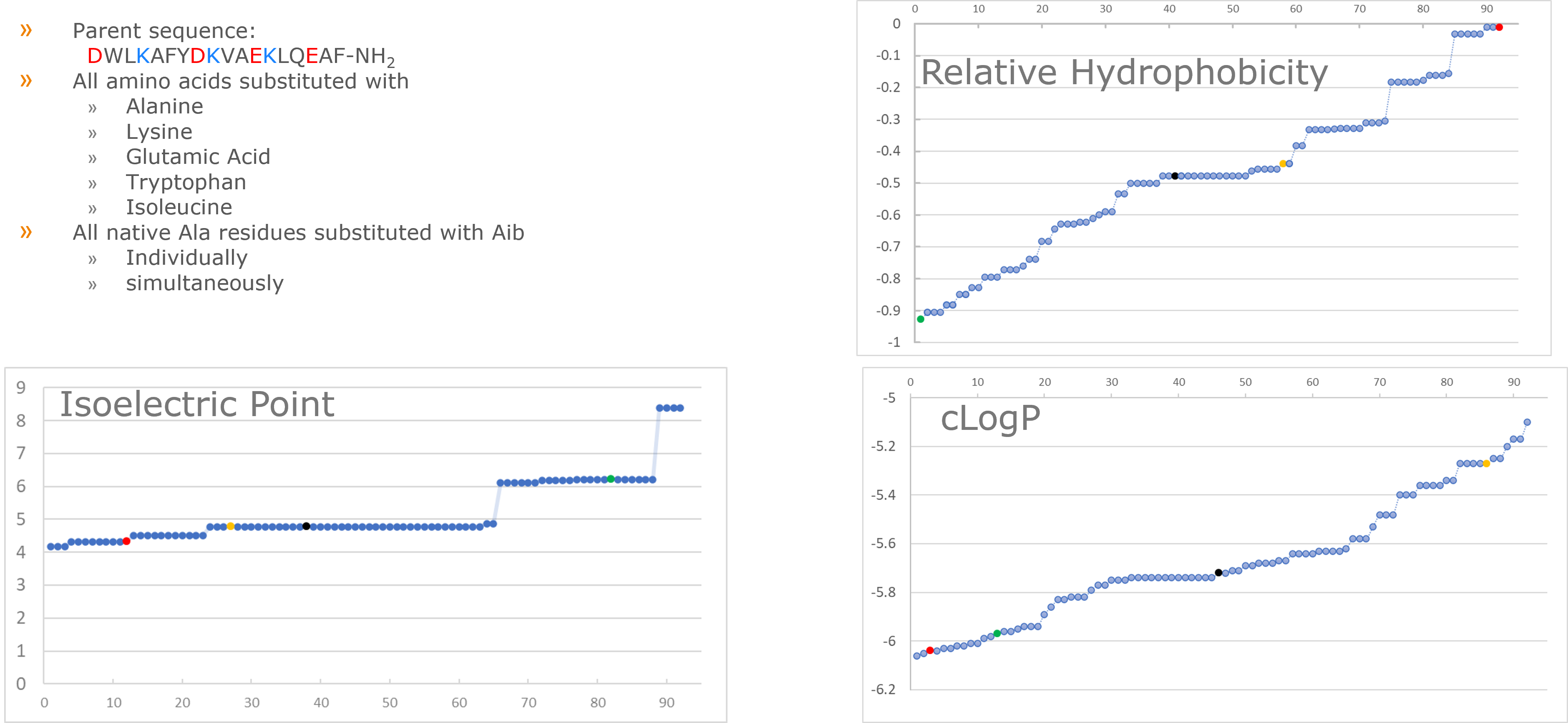
Figure 1. Description of synthetic peptide library under investigation and properties of resulting library members. Red circle represents the most hydrophobic peptide (K4I), yellow circle represents a median hydrophobicity (L14I), green represents the most hydrophilic sequence (V10K) and black circle represents the parent peptide sequence on the charts for various physiochemical properties.
I like this peptide as a "parent" sequence for a couple of reasons:
- -the linear sequence is relatively straightforward to synthesize as an individual
- -the parent peptide sequence is of similar size to those currently being investigated for therapeutic applications.
- -the peptide has sufficient length to fill out a full 96-compound library using amino acid scanning
- -amino acid scanning will introduce a wide variety of sequence diversity, which it turn could correlate with synthesis difficulty
To synthesize this library, I decided to use my Biotage® Syro II in the Tip Synthesis configuration. Tip synthesis allows for synthesis of peptides on 1-5 µmol scale using a 0.4 mL fritted pipette tip for reactor vials. Given the small liquid volumes used in these reactor vials, the vials are stationary. To ensure synthesis success then, double coupling and amino acid preactivation are employed.
I knew that I would use 5 equivalents and 0.5 M amino acid solutions, but at this point I decided to think a bit harder about the specific coupling reagents used for this library. Typically, I'm one to choose one reagent and stick with it throughout the whole synthesis. It simplifies programming of the synthesizer and reduces time spent preparing reagents. But I recalled a publication in which two different coupling reagents were used with greater success for synthesis of aggregation prone sequences. Using this information as a starting point, I decided to synthesize the library using first DIC/OxymaPure then HATU/DIPEA for the second coupling. In a second synthesis, I then switched the order of coupling reagents and looked to see if there were any significant differences in crude purity.
Don't worry - I'm not going to share with you 192 different HPLC chromatograms - but I would like to focus on the sequences highlighted previously, Figure 2.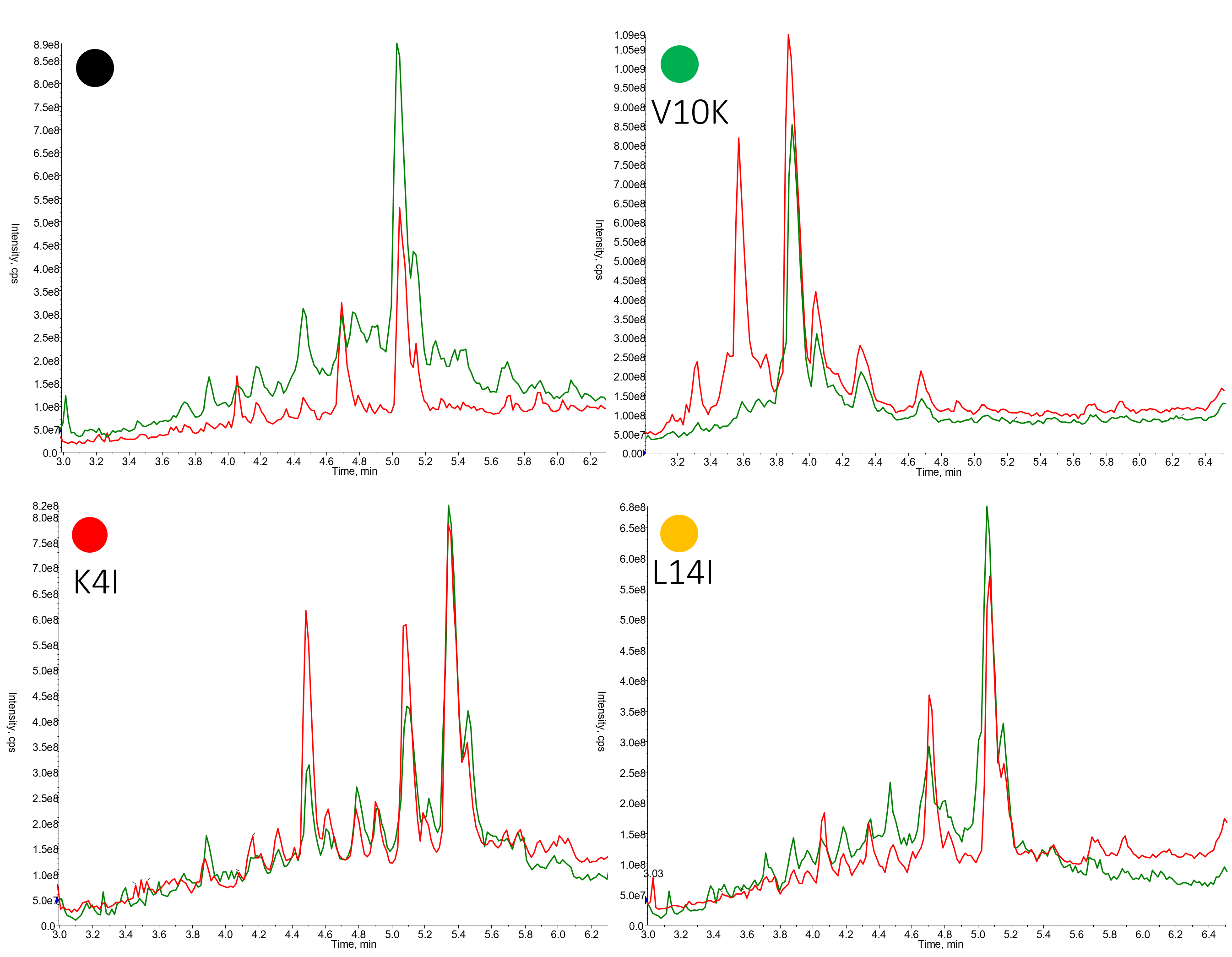
Figure 2. Representative crude chromatograms for members of the 18A peptide library synthesized using DIC/OxymaPure followed by HATU/DIPEA (red trace) or HATU/DIPEA followed by DIC/OxymaPure (green trace).
For the peptides shown here, the protocol using HATU/DIPEA followed by DIC/OxymaPure generally produced peptides with higher crude purity. What I find most interesting though is just how different the two chromatograms are in some cases (V10K variant for example). Simply switching the order that the two different coupling reagents are used eliminates completely the presence of a significant impurity. On the other hand, some crude purity profiles are strikingly similar (K4I variant).
The differences are less surprising, given just how different the activated amino acid species are when compared to each other. Not only are they significantly different structurally, Figure 3, but they also have different reaction kinetics of formation, condensation, and lifetime, which all impact overall coupling efficiency.
Figure 3. Structure of a generic amino acid activated using either DIC/OxymaPure (left) or HATU/DIPEA (right).
As with any peptide library, there were a couple sequences that stood out as particularly terrible from a synthetic perspective, Figure 4. 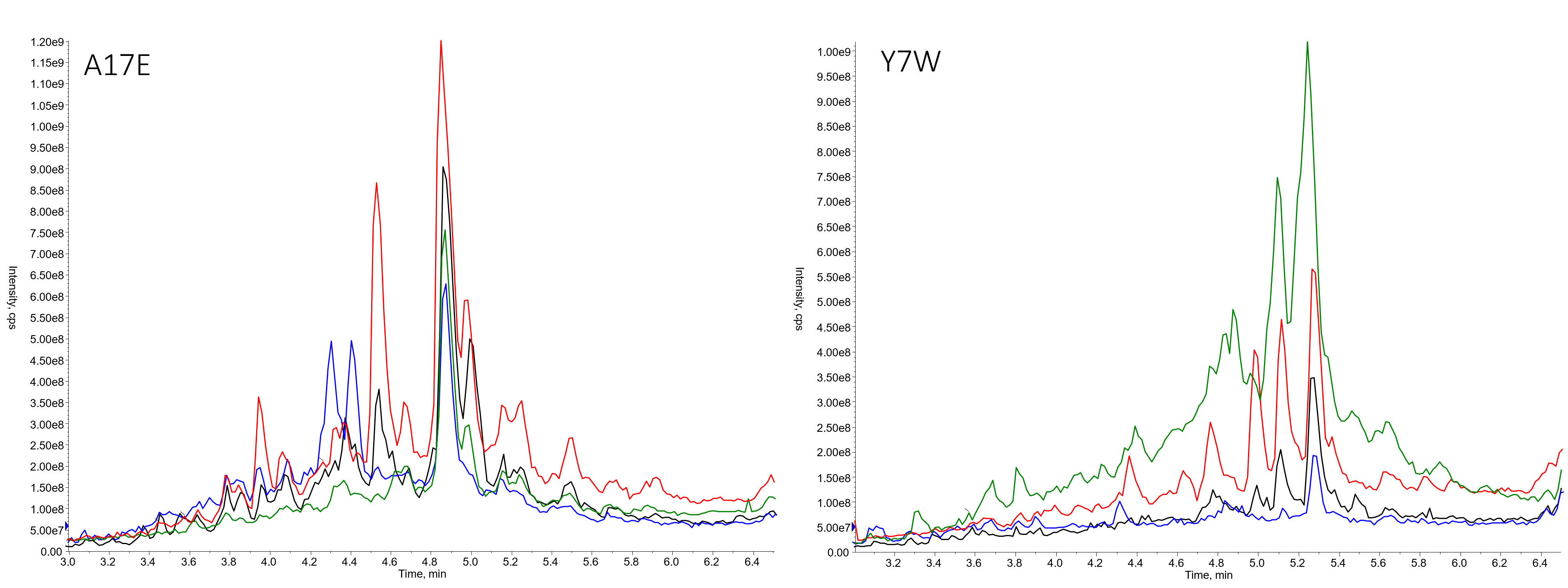
Figure 4. Library members prepared using DIC/OxymaPure then DIC/OxymaPure (black), HATU/DIPEA then HATU/DIPEA (blue), DIC/OxymaPure then HATU/DIPEA (red) or HATU/DIPEA or DIC/OxymaPure (green).
These results indicate that there probably won't be a tried and true, use this protocol for all libraries rule. This isn't unexpected, given the synthetic diversity that could be present in a single library. What I would like to highlight though is that the mixed coupling reagent protocol, specifically using HATU/DIPEA for the first coupling reaction and DIC/OxymaPure for the second coupling reaction gave the best results for the A17E library variant, adding further evidence my hypothesis that two different coupling reagents can in fact improve the crude purity of the library as a whole.
What is clearly evident in these data, and others not shown, is that the use of two difference coupling reagents for synthesis of peptide libraries at particularly small scale improves the overall crude purity of the library. Also, the order in which two different coupling reagents are used during peptide synthesis will, unsurprisingly, impact the synthesis outcome.
If you would like to learn about more strategies for synthesis of peptide libraries, click the link below and achieve even greater crude purities.

