Jan 10, 2024 3:00:00 PM
Does C-terminal amino acid affect cyclization efficiency?
By Austin Schlirf

Head-to-tail cyclization of peptides is being used in drug development as yet another way to probe how constraining peptide sequence structure in solution can improve pharmacokinetics and target specificity. Head-to-tail cyclization on resin has been seen as difficult and low yielding; however, I have explored several ways to improve or at least approach head-to-tail macrocycles to make this process more efficient. In this post, I will explore how the identity of the C-terminal amino acid can affect this efficiency.
When I started this journey, I had originally attempted to compare Asp and Glu as my C-terminal amino acids that linked to the resin via their side chains. For my 15-mers I had several side products and low purity for using Asp as a linker and I could not figure out why. Granted, this was my first experience with macrocycles so, on top of synthesis, I was also learning a bit about cyclic peptide workflow; this just goes to show that improvement doesn’t happen overnight. Subsequently I went on to explore what peptide length and resin loading did to improve the synthesis and now it is time to see if the Asp linked peptide can be synthesized with improved purity and yield and compare that to the Glu linked peptide.
I decided to use my 10-mer sequence, AWELKRDVAD, as I had better linear purity compared to the 15-mers, and improved handling compared to the 5-mers, which really liked to dimerize. So, I synthesized my 10-mer Asp-linked peptide at a 0.125 mmol scale using a Biotage® Initiator+ Alstra™ peptide synthesizer using standard DIC/Oxyma coupling methods. Below is my linear purity compared to my Glu-linked linear peptide, Figure 1.
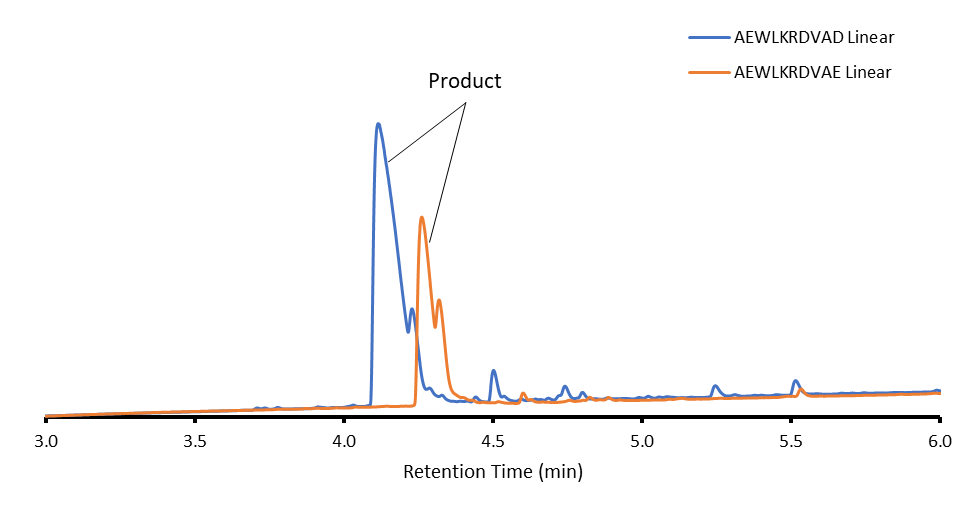
Figure 1: Chromatograms of linear products between Glu linked and Asp linked 10-mers.
So, as I saw with my Glu linked peptide, my Asp sample is way purer compared to its 15-mer counterpart probably due to the nature this sequence which is why I chose to compare the 10-mer samples. From here I cyclized both samples using my standard DIC/Oxyma double coupling procedure. Below we can see the final purity of both products, Figure 2.
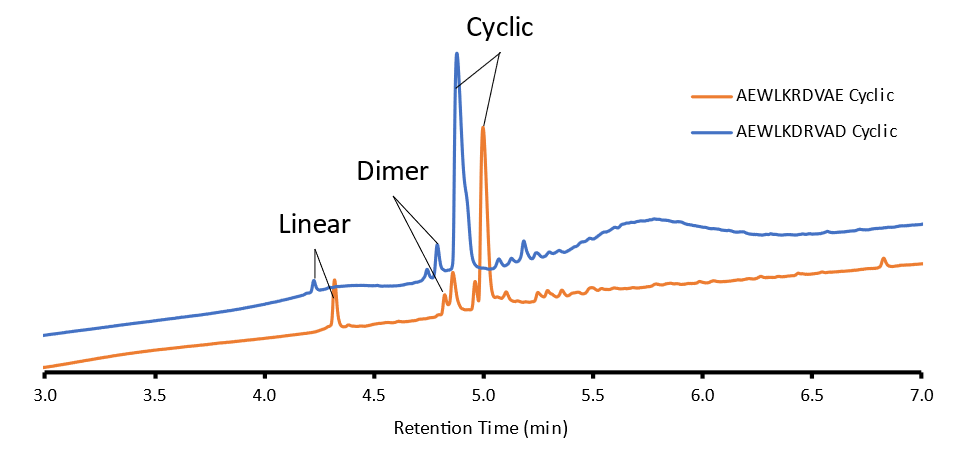
Figure 2: Chromatogram of Glu and Asp linked cyclic peptides.
The crude purity of the Asp-linked cyclic peptide was 74.7% with 1.3% linear product and 9.92% dimer. The crude purity of the Glu-linked cyclic peptide was 52.5% with 8.18% linear and 19.46% dimer. Here we see that there is significantly more linear impurity and dimer from the Glu-linked peptide compared to the Asp-linked peptide. When I was first trying to figure out the possibility for head-to-tail cyclization on resin, the Asp-linked 15-mer was much less successful than the Glu-linked peptide. I initially thought the Glu peptide was more efficient at cyclization due to the extra degrees of freedom an extra carbon provided in the side chain. This flexibility would lead to better cyclization efficiency because the peptide could fold back and react with itself at the resin surface more easily. Here however we can see that the Asp may aid in reducing dimerization in shorter peptides leading to higher purity— this is possibly due to reduced flexibility. The N-terminus is more localized to its origin active site and therefore is more likely to find its C-terminus rather than a neighbor’s. This coupled with a shorter chain promotes intramolecular cyclization rather than intermolecular cyclization. Honestly, this collective suite of data also demonstrates that not every strategy works for every peptide, which is something I have been learning on my peptide journey.
If you want to learn more about automating cyclic peptide synthesis, check out the link to our application note below and let us know if you have any questions or want to talk strategies for your syntheses!
Published: Jan 10, 2024 3:00:00 PM


