Jan 30, 2023 12:43:37 PM
Analyzing crude peptide samples by Mass Spectrometry: what are the options
By Elizabeth Denton

There are many techniques available to analyze and identify synthetic compounds that we are taught in the first few years of our chemistry education. While tools like NMR spectroscopy, IR spectroscopy, and others, are extremely useful for determining or confirming the structure of synthetic small molecules, these strategies are not as well suited for quick characterization of peptides. As a result, peptide chemists rely heavily on peak shape observed during a liquid chromatography step and mass spectrometry for mass confirmation.
In today's post, I'll discuss several of the mass spectrometry techniques that are used for analyzing crude or purified peptide samples.
Before we get too far into this, I just want to say that I am by no means an expert in mass spectrometry (MS) and all the unique features of each particular strategy but I hope to provide a more general overview of some of the ionization strategies currently used. That being said, I think that MS and the advances made in this space are awesome and I only expect more in the future!
To be detected by a mass spectrometer, a compound must first be ionized (either positively or negatively charged). The most commonly used ionization techniques are:
- -Matrix-Assisted Laser Desorption Ionization (MALDI)
- -Electrospray Ionization (ESI)
- -Chemical Ionization (CI)
- -Atmospheric Pressure Chemical Ionization (APCI)
- -Electron Impact Ionization (EI)
- -Fast Atom Bombardment (FAB)
However, for many of these ionization methods, applications in the peptide or protein space simply aren't reasonable. Methods like APCI, EI and FAB are often much too aggressive for analyzing peptides as they often result in fragmentation of the compounds of interest. While this could be advantageous if fragmentation was predictable, however, for a simple Yes/No answer, the generated spectra are much too complicated.
Therefore, ESI and MALDI now dominate the mass spectrometry field as the ionization methods of choice. These strategies are gentle enough that ionization, detection, or even sequencing of intact peptides (or their multiply-charged ion states), proteins, and more recently, intact protein complexes are possible. So how do these methods work, and why are they so well suited to bio-polymers?
In MALDI MS, a sample is diluted into a suitable matrix solution, spotted onto a plate, allowed to dry, and then ionized into the vapor phase using laser light.
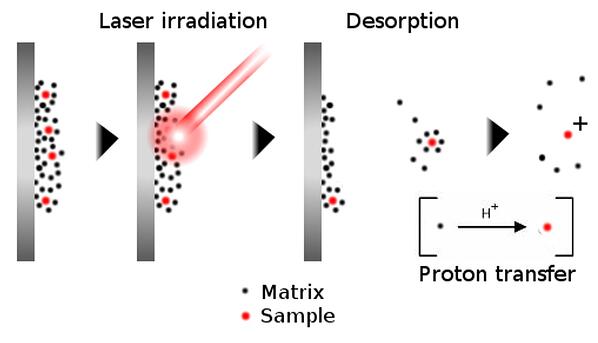
Figure 1. Principle of ion generation using MALDI mass specrometry. A non-volatile sample is diluted into a matrix solution, which dries onto a plate. Striking the sample with a laser vaporizes the matrix-sample solid, whereby proton transfer occurs, ionizing the sample for detection.
The matrix, of which there are many varieties, serves several primary roles:
-the matrix facilitates vaporization of compounds that are not commonly volatile - think peptides and proteins
-matrix compounds act an alternative to absorb some of the laser energy, limiting fragmentation of the compound of interest
-the matrix serves as a chemical source of ions that ionize the compound of interest once vaporized
Because the matrix itself contributes protons to ionization, the compounds detected using this technique most often only carry a single charge - allowing rapid identification of the parent compound. Importantly, the samples are spotted onto a MALDI plate before analysis, therefore MALDI-MS can also be considered pseudo-high throughput. The MALDI plates I have used in the past have 96 spots and analyzing each spot takes only a minute or so. I used MALDI most often to identify the peaks of a scouting run, especially for the first time synthesis of a peptide.
During electrospray ionization, the compound is introduced into the source in a liquid solution and then the solution is sprayed out of a capillary, directed with a heated gas stream towards the cone, assisted by an electrical current, and into the mass detector, Figure 2. Because of the demand for a carrier solvent for introducing the sample into the source, ESI is uniquely suited to directly connect a liquid chromatography system to a wide variety of mass detectors (a discussion for later).
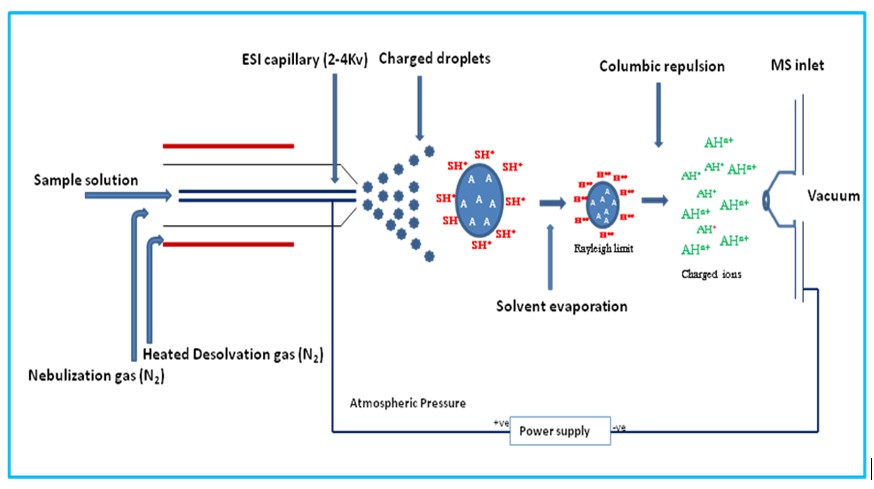
Figure 2. Principle of ion generation using an electrospray ionization source. The sample ions are formed in solution with a carrier solvent then sprayed through a capillary into the ion source and directed towards the detector cone with a heated carrier gas and an electrical current. While translocating through the source, the carrier solvent droplets evaporate, leaving behind only the sample ions for introduction into the detector.
During the translocation, the droplet containing the sample evaporates, leaving behind the ionized compound for introduction into the mass detector. Through this process, compounds can take on multiple protons, yielding charged compounds with multiple positive charges (+2, +3, +4, and up are common for peptides!). While this seems like it could increase the difficulty of the analysis, mass spectrometry software has improved such that deconvolution packages are now available that can recombine each multiply-charged variant of the desired peptide up to the parent mass. Importantly though, multiply-charged ions bring the detected mass/charge down into within most mass spectrometers' detection range (often 2000-3000 Da). Remember, in mass spectrometry the detected mass is not a true mass, but a ratio of mass per charge (m/z).
Now that we've created ions and detected their presence. How do you know what you're looking at? For MALDI ionization, the answer is clear. The mass species detected is more often than not a singly charged, parent mass ion. No extra work necessary!
For ESI methods though, a little more thought it necessary. For instruments with lower sensitivity, you'll likely only see the major mass with a Gaussian peak shape (ideally). In this case, the deconvolution software I mentioned earlier will certainly simplify your life. Luckily though, since you've synthesized this peptide, so you should know what the expected molecular weight is of the final compound. To figure out what the masses for the multiply charged ions should be, use the formula
(MW + x)/x
where x is the number of protons added that contribute to the overall charge state of the peptide. For example, to calculate the mass of a peptide in a +3 charge state simply add 3 Da to the peptide molecular weight then divide this number by three.
If you happen to be using a high resolution mass detector, you can often resolve not only the predominant mass, but also each mass in proportion to the isotopic abundance of the different atom isotopes (C13, N15, etc), Figure 3. Taking a closer look at the mass difference between each of these peaks will tell you to which charge state that specific peak correlates.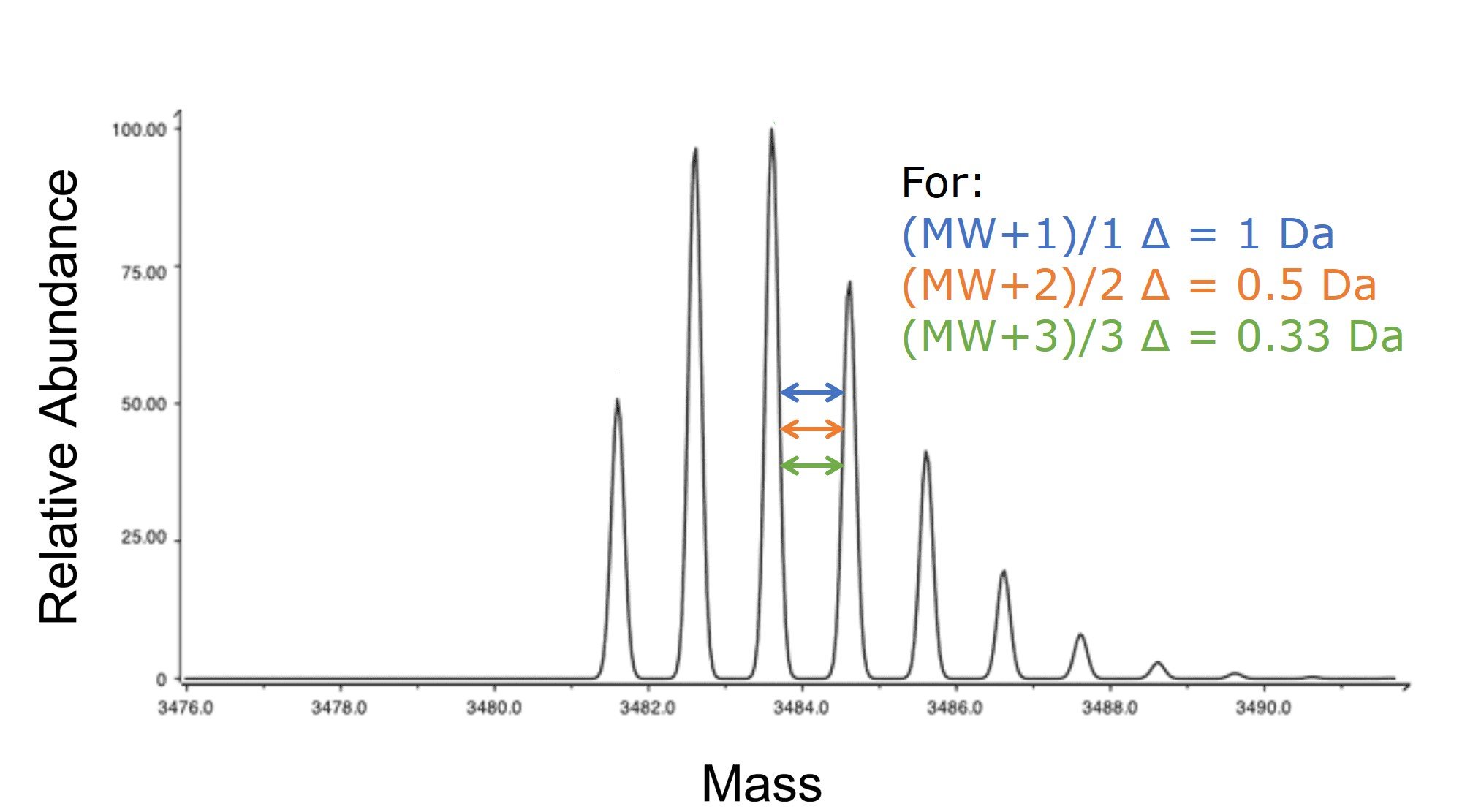 Figure 3. Isotopic mass envelope generated with an ESI source and detected by a mass spectrometer with high sensitivity. The difference in mass between the peaks are indicative of the charge state measured with the specific mass of interest.
Figure 3. Isotopic mass envelope generated with an ESI source and detected by a mass spectrometer with high sensitivity. The difference in mass between the peaks are indicative of the charge state measured with the specific mass of interest.
Because mass spectrometry has become such a critical tool for peptide chemists characterizing their synthetic peptides, it is important to have a working understanding of what is going on inside that big expensive box. Different ionization strategies can yield different results, and I hope this post has served to clarify the significant difference between the most common methods used in the peptide space today.
To learn more about how Biotage tools can improve your peptide synthesis and overall workflow efficiency, follow the link below.
Published: Jan 30, 2023 12:43:37 PM

